Quick Look
Grade Level: 10 (9-12)
Time Required: 1 hours 15 minutes
Lesson Dependency:
Subject Areas: Biology
Summary
The discovery of restriction enzymes and their applications in DNA analysis has proven to be essential for biologists and chemists. This lesson focuses on restriction enzymes and their applications to DNA analysis and DNA fingerprinting. Use this lesson and its associated activity in conjunction with biology lessons on DNA analysis and DNA replication.Engineering Connection
Microfluidics concepts and devices used to study colloidal particle flow are employed by biologists to study and filter biomolecules. Gel electrophoresis is one example of the many and creative engineering applications that are used by scientists to compare fragments of DNA samples.
Learning Objectives
After this lesson, students should be able to:
- Describe the basic steps involved in DNA fingerprinting.
- Explain the function of restriction enzymes and the role of restriction sites along the DNA.
- Explain how DNA fragments are charged molecules.
- Explain that the distance traveled by DNA fragments inside a gel tray depends on fragment length
- Explain that the DNA (genetic code) from related organisms contain similar restriction sites and related DNA are cut into similar fragments by the same restrictions enzymes.
Educational Standards
Each Teach Engineering lesson or activity is correlated to one or more K-12 science,
technology, engineering or math (STEM) educational standards.
All 100,000+ K-12 STEM standards covered in Teach Engineering are collected, maintained and packaged by the Achievement Standards Network (ASN),
a project of D2L (www.achievementstandards.org).
In the ASN, standards are hierarchically structured: first by source; e.g., by state; within source by type; e.g., science or mathematics;
within type by subtype, then by grade, etc.
Each Teach Engineering lesson or activity is correlated to one or more K-12 science, technology, engineering or math (STEM) educational standards.
All 100,000+ K-12 STEM standards covered in Teach Engineering are collected, maintained and packaged by the Achievement Standards Network (ASN), a project of D2L (www.achievementstandards.org).
In the ASN, standards are hierarchically structured: first by source; e.g., by state; within source by type; e.g., science or mathematics; within type by subtype, then by grade, etc.
International Technology and Engineering Educators Association - Technology
-
The sciences of biochemistry and molecular biology have made it possible to manipulate the genetic information found in living creatures.
(Grades
9 -
12)
More Details
Do you agree with this alignment?
-
Connect technological progress to the advancement of other areas of knowledge and vice versa.
(Grades
9 -
12)
More Details
Do you agree with this alignment?
State Standards
Texas - Science
-
know that hypotheses are tentative and testable statements that must be capable of being supported or not supported by observational evidence. Hypotheses of durable explanatory power which have been tested over a wide variety of conditions are incorporated into theories;
(Grades
9 -
11)
More Details
Do you agree with this alignment?
-
know scientific theories are based on natural and physical phenomena and are capable of being tested by multiple independent researchers. Unlike hypotheses, scientific theories are well-established and highly-reliable explanations, but they may be subject to change as new areas of science and new technologies are developed;
(Grades
9 -
11)
More Details
Do you agree with this alignment?
-
distinguish between scientific hypotheses and scientific theories;
(Grades
9 -
11)
More Details
Do you agree with this alignment?
-
plan and implement descriptive, comparative, and experimental investigations, including asking questions, formulating testable hypotheses, and selecting equipment and technology;
(Grades
9 -
11)
More Details
Do you agree with this alignment?
-
analyze the levels of organization in biological systems and relate the levels to each other and to the whole system.
(Grades
9 -
11)
More Details
Do you agree with this alignment?
Pre-Req Knowledge
Basic human anatomy, high school freshman-level biology.
Introduction/Motivation
How do scientists compare DNA samples? What tools do they use? Comparison between multiple DNA samples can be performed by a simple technique called restriction fragment length polymorphism (RFLP). This technique is based on several concepts ranging from genetics to physics and engineering. The genetic code contained inside a DNA molecule does not need to be entirely compared between two or more DNA samples, only some portions of it.
In the early days of genetics research, no computers or automated and speedy processes for DNA decoding and analysis existed. So how was it possible to analyze and compare DNA samples? By analyzing short DNA fragments! Restriction enzymes are a special class of enzymes that can cut the DNA into fragments at specific locations called restriction sites. This is a defense mechanism employed by bacteria for protection against viral DNA or genetic code. Researchers realized that this behavior could be exploited to cut DNA into short fragments for lab analysis.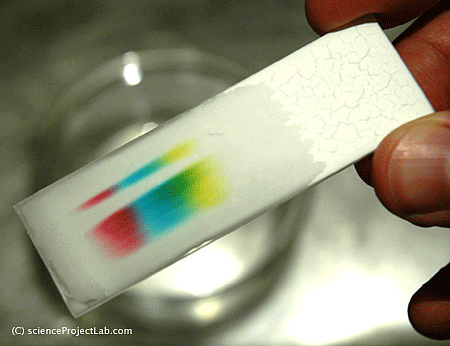
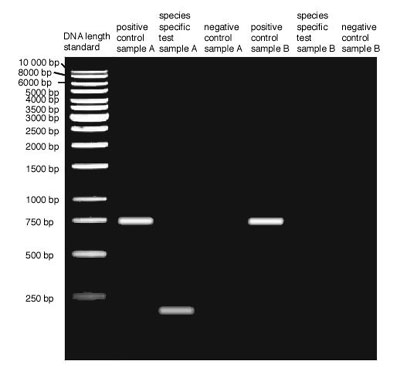
How can multiple DNA samples be compared? By cutting them with the same restriction enzyme and comparing the fragments. If two or more DNA samples have the same restriction sites, then they will be broken into similar fragments. How can someone tell if two DNA fragments are the same? This is accomplished by placing the fragments in a gel tray and applying an electric current along the plate. As the DNA fragments are charged, the positively charged fragments move toward the cathode and the negatively charged toward the anode. How fragments move through the agarose gel solution is related to their masses—smaller fragments move further away faster, while longer fragments move slower.
A similar example to this process is how the pigments that are used to give a certain color to paint segregate when a drop of water-based paint is placed on a filter paper and water is allowed to diffuse through the paper (this phenomenon is illustrated in Figure 1). Through this process, paint colors that are the result of combinations of multiple pigments decompose into their respective pigments, thus, allowing for the identification of common pigments.
Lesson Background and Concepts for Teachers
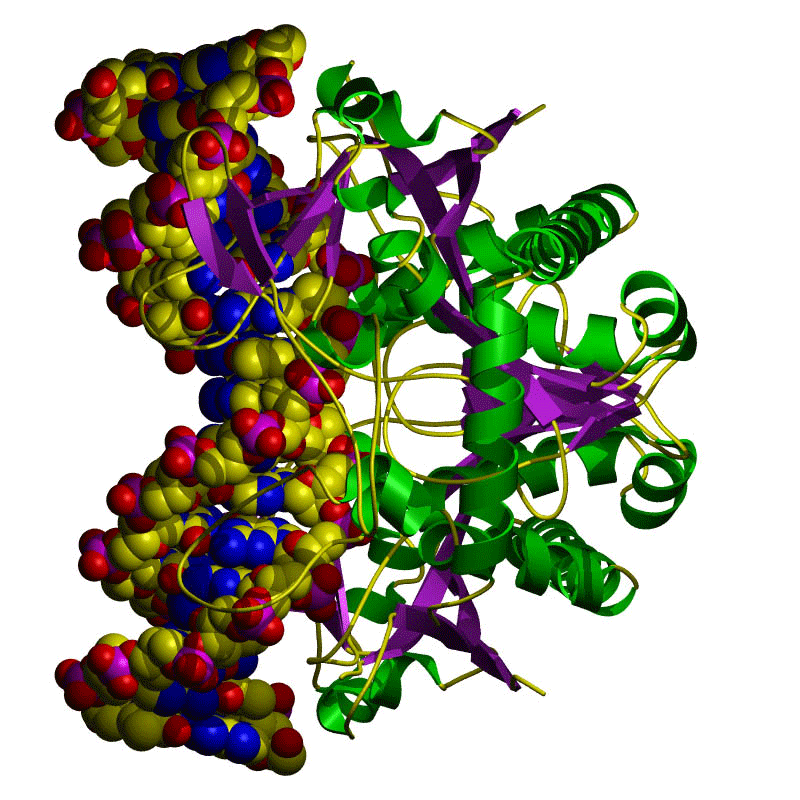
Restriction enzymes attach to DNA and are activated by restriction sequences in the DNA. Once activated, the restriction enzymes hydrolyze and destroy the bonds between nucleotides. The restriction sequences along the DNA are inherited, thus, people who are related have similar restriction sequences along their DNA. Cutting DNA samples by the same restriction enzymes and analyzing the resulting DNA fragments by DNA fingerprinting indicates which DNA samples have similar restriction sequences.
During DNA fingerprinting, fragments are placed in agar gel and an electric field is applied along the gel plate. As DNA fragments are electrically charged, they travel through the gel. DNA fragments of the same length travel at the same rate, with shorter fragments traveling faster and longer than slower ones. DNA samples that are broken in similar fragments have the same restriction sequences occurring at the same locations along the DNA double strand. Students can conduct the fun and hands-on associated activity DNA Forensics and Color Pigments to perform DNA forensics on liquid food coloring (water-based colors) to enhance their understanding of DNA fingerprinting,
Restriction enzymes attach to DNA and cleave it (cut it) randomly or at specific locations. Bacteria are protected from foreign DNA by using restriction enzymes to destroy the foreign DNA. Restriction enzyme (restriction endonuclease) cuts DNA at specific locations (specific nucleotide sequence) called restriction (recognition) sites. But how is the DNA of the organism (or bacteria) protected against its own restriction enzymes? The DNA is protected by methylases—enzymes that add methyl groups to adenine and cytosine that are part of the recognition sequences (sites) to prevent the restriction enzymes from cleaving the DNA. Restriction enzymes are categorized in:
- Type I and III: both restriction and methylation performed by one large enzyme
- Type II and III: cleave (cut) DNA at random sites (do not need recognition sequence)
- Also, Type II—only restriction, methylase performed by another enzyme.
- Examples of restriction enzyme: EcoRI. It cleaves at the GAATTC recognition sequence. The DNA double helix is cut between G and A and two "sticky ends." 5' (AATT) result. Sticky ends can bind to their complement (DNA ligase enzymes fuse two fragments—just the opposite action of restriction enzymes).
Associated Activities
- DNA Forensics and Color Pigments - Students perform DNA forensics on liquid food coloring (water-based colors) to enhance their understanding of DNA fingerprinting, restriction enzymes, genotyping and DNA gel electrophoresis.
Assessment
As a summary assessment for this lesson and its associated activity, have students complete the DNA Forensics and Color Pigments Worksheet attached to the activity. Review student answers to gauge their comprehension of the material presented.
Subscribe
Get the inside scoop on all things Teach Engineering such as new site features, curriculum updates, video releases, and more by signing up for our newsletter!More Curriculum Like This
Students perform DNA forensics using food coloring to enhance their understanding of DNA fingerprinting, restriction enzymes, genotyping and DNA gel electrophoresis.
Students are introduced to the latest imaging methods used to visualize molecular structures and the method of electrophoresis that is used to identify and compare genetic code (DNA).
Students learn how engineers apply their understanding of DNA to manipulate specific genes to produce desired traits, and how engineers have used this practice to address current problems facing humanity. Students fill out a flow chart to list the methods to modify genes to create GMOs and example a...
Copyright
© 2013 by Regents of the University of Colorado; original © 2012 University of HoustonContributors
Mircea Ionescu; Myla Van DuynSupporting Program
National Science Foundation GK-12 and Research Experience for Teachers (RET) Programs, University of HoustonAcknowledgements
This digital library content was developed by the University of Houston's College of Engineering under National Science Foundation GK-12 grant number DGE-0840889. However, these contents do not necessarily represent the policies of the NSF and you should not assume endorsement by the federal government.
Last modified: July 1, 2019





User Comments & Tips